Abstract
Despite the effectiveness of tyrosine kinase inhibitors (TKIs) against chronic myeloid leukemia (CML), acquired drug resistance often leads to relapse. CML has unique microRNA (miRNA) expression profiles at different disease stages and in response to TKI-treatment. However, the predictive utility of miRNAs is not yet conclusive and there is a need to identify key biomarkers for prognostic tools to be used in the clinic. We have recently generated global transcriptome profiles on treatment-naïve CD34+ CML cells with known subsequent imatinib (IM) responses and identified several differentially expressed miRNAs, including miR-185 and miR-145, in IM-nonresponders as compared to IM-responders. We also reported that IM response could be predicted in treatment-naïve CD34+ cells by an in vitro colony forming cell (CFC) assay.
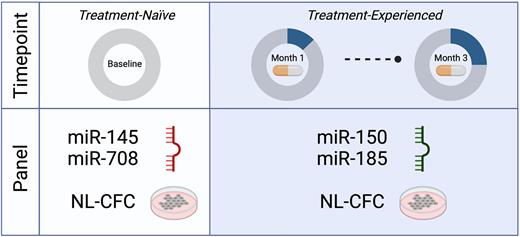
In this study, we evaluated miRNA expression changes in CD34+ CML cells pre- and post-nilotinib (NL) therapy from 58 CML patients enrolled in the Canadian sub-analysis of the ENESTxtnd phase IIIb clinical trial and assessed a potential correlation between miRNA expression and sensitivity of CD34+ cells to TKIs in CFC assays, to predict NL response in these patients. CFC assays were performed by plating pre-treated CD34+ cells with methylcellulose-medium ± Imatinib (IM), NL and Dasatinib (DA) and showed that only NL was able to differentiate responses between NL-responders and NL-nonresponders (p=0.011) and predict NL response (p=0.00016) based on calculated cutoffs. Microfluidic qRT-PCR and a univariate Cox proportional hazard (CoxPH) analysis on miRNA expression profiles were then performed in CD34+cells obtained at diagnosis (BL), 1-month (M1) and 3-month post-NL treatment (M3) from 58 CML patients; this showed that 17 out of 47 miRNAs examined were significantly associated with NL response (p<0.05). Further Welch t-test analysis revealed that nine of these miRNAs were differentially expressed between NL-responders and NL-nonresponders. These miRNA candidates were then subject to a multivariate CoxPH analysis ± CFC assay data from each patient, which demonstrated that miR-145 (p=0.013) and miR-708 (p=0.009) together could improve predictive power at the BL state. Additionally, receiver-operating-characteristic (ROC) and precision-recall (PR) area-under-curve (AUC) values (1.2-fold) of performance plots generated by random forest (RF) and Naïve-Bayes (NB) machine learning algorithms were increased in this multivariate panel compared to individual miRNA variables. At M1 and M3, four miRNAs were found to be stably associated with NL response individually and a combination of miR-150 (M1 p<0.001, M3 p=0.01) and miR-185 (M1 p=0.009, M3 p=0.01) was significantly associated with treatment response in multivariate CoxPH analysis. Again, the multivariate analysis improved ROC and PR AUC values, especially for miR-185 (1.2 to 2-fold). Most interestingly, incorporation of NL-CFC output consistently increased AUC values up to 2-fold at both BL and M1/M3 time points that enhanced predictive performance.
Together, these findings offer two predictive models for NL response in treatment-naïve or post-treatment CML patients, which could be developed into prognostic biomarkers.
Disclosures
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.